Research
The research in the Taipale lab is organized around two central themes: technology development in functional genomics and proteomics and the organization and function of the human protein homeostasis network in health and disease. Individual projects in the lab are very diverse, spanning bacterial toxins to transcriptional regulation to rare disorders to chaperone/client interactions. But despite this diversity, all projects share common features. We take advantage of – or develop new – large-scale systematic ‘omics’ methods and use them as an entry point to understand biological processes. In particular, we aim to understand these processes through the lens of interactions between molecules – be they proteins, DNA, RNA, or small-molecule compounds. Another similarity is that all projects involve close collaborations with other research groups. Not only are collaborations a lot of fun, but they also allow us to tackle the inevitable tradeoff between the breadth and depth of expertise.
Technology development
Technology development is one of the main drivers of modern biomedical research. For example, RNA interference, cellular reprogramming, CRISPR/Cas9, next-generation (single-cell) sequencing, and large-scale DNA synthesis have transformed research in the last 20 years, and few would doubt that that future technologies will be similarly impactful. We are focused on developing new technologies to identify biologically and therapeutically important biomolecular interactions, and to find ways to facilitate translating such interactions to functional insights and therapeutic interventions.
Ligand/receptor interactions. Cells secrete thousands of diverse proteins, collectively known as the secretome. The secretome plays a fundamental role in development, growth control, tissue homeostasis, cell-cell communication, and immunity. Modern protein/protein interaction assays have been very successful in characterizing stable interactions between soluble intracellular proteins. However, there are few scalable methods for studying extracellular interactions, although over 60% of FDA-approved drugs target extracellular proteins. We are developing methods to tackle this challenge by harnessing the power of highly potent bacterial and plant exotoxins. Our aim is to systematically de-orphanize secreted proteins in the human genome.
We are also interested in the interactions between exotoxins and their receptors. We recently identified semaphorins SEMA6A and 6B as the receptors for P. sordellii Lethal Toxin TcsL and, in collaboration with the Julien and Melnyk labs, characterized the molecular basis for receptor specificity among this toxin family. We are continuing to characterize how exotoxins evolve new receptor specificities and how this could be exploited for therapeutic purposes.
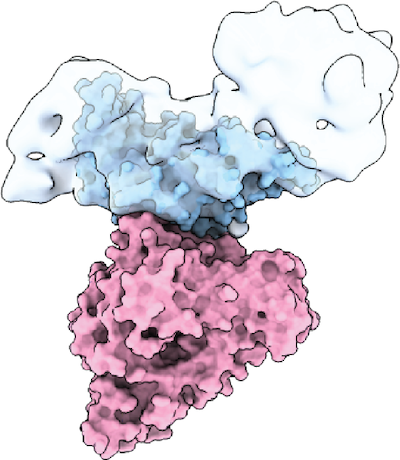
TcsL (blue) bound to SEMA6A (pink) (Lee et al. Cell 2020)
“Instead of discovering which molecules interact with each other, we force molecules to interact with each other and see what happens.”
Induced proximity proteomics. Modern proteomics and genomics approaches are often based on the guilt-by-association principle. If an unknown gene physically, genetically, or computationally associates with a known gene, the function of the unknown gene is inferred from the known partner. This has proven a very successful principle in ‘omics’ research. However, guilt-by-association does not always work, since association does not imply function. To address this challenge, we have turned the tables. Instead of discovering which molecules interact with each other, we force molecules to interact with each other and see what happens. Such “induced proximity proteomics” allows us to discover proteins based on function rather than association. As an example, using this approach we have identified hundreds of novel proteins that can activate or repress transcription when tethered to genomic loci. We are applying similar methods to other central cellular processes, such as protein homeostasis.
Protein homeostasis
It’s tough to be a protein. From the moment of birth in the ribosomal exit tunnel until the very last squeeze through the proteasomal channel, they are often harassed by various stresses. To cope with this, cells have evolved an elaborate network of proteins that maintain protein homeostasis, or proteostasis. This evolutionary ancient network consists of chaperones and co-chaperones, the ubiquitin-proteasome system, trafficking proteins, and hundreds of other quality-control factors. In turn, these factors associate with and regulate essentially all cellular proteins. We are interested in deciphering how this network is physically organized in human cells. We use diverse high-throughput protein/protein interaction assays and CRISPR/Cas9 screens to characterize the network and identify the key players.
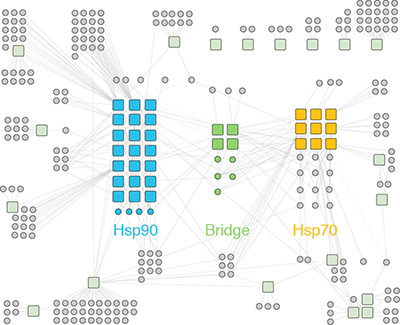
Organization of the human protein homeostasis network. We are particularly interested in how a limited set of quality control factors regulate the rest of the proteome. For example, the Hsp70 and Hsp90 chaperones promote the folding and function of the majority of the proteome. To do so, these major players associate with an oddball cast of diverse co-chaperones that determine the client specificity. The evolution of more complex proteomes has coincided with the expansion of co-chaperone families. For example, the human genome encodes 47 different Hsp70 co-chaperones called J domain proteins (or Hsp40s). But the function of most J domain proteins is unknown. We are studying the human Hsp70/J protein/client network with a panel of sensitive protein/protein interaction assays, and functionally characterizing novel roles of J domain proteins in the cell. Beyond chaperones, we are also interested in studying other abundant proteins that interact with hundreds if not thousands of clients, such as 14-3-3 proteins.
Similarly, we are interested in how E3 ligases and deubiquitinases (DUBs) regulate the function and stability of thousands of proteins. We are using functional genomics tools, including CRISPR/Cas9 screens, to identify E3 ligases and DUBs that regulate the function of therapeutically relevant target proteins such as oncogenes and tumor suppressors.
rare diseases
%
with treatments
Can we do better?
Systematic characterization of pathogenic disease variants. There are about 7,000 known Mendelian diseases. While each of them is rare, together they affect millions of people around the world. We already know the genes underlying Mendelian disorders. But what then? There is no cure nor treatment for most of these diseases, and pharmaceutical companies have no financial incentives to develop drugs for them.
The traditional approach has been to use multiple assays and technologies to study a particular Mendelian disease. But what if we turned the question upside down? What if we studied many rare diseases at the same time but with very specific assays? Although these diseases are incredibly diverse and affect different pathways and tissues, there are many commonalities between them too. For example, missense mutations, which account for about half of all Mendelian disorders, often do not lead to simple loss of protein function. Rather, they disrupt protein function in more subtle ways by tuning protein folding pathways, altering subcellular trafficking, interrupting signaling cascades, and rewiring highly connected interaction networks. We take advantage of these commonalities between pathogenic variants and systematically characterize the effect of pathogenic variation on protein/protein interactions, protein trafficking and other phenotypes. For example, we have characterized the subcellular localization of over 4,000 pathogenic disease variants involved in more than 1,000 Mendelian disorders.
In the long run, we want to use these pathogenic phenotypes to systematically identify pathways and drugs that could restore the function of any of these mutant proteins. In other words, instead of looking for a drug for a disease, we want to find a disease for a drug.
